down-mx.img.susercontent.com/file/cn-11134207-7r98
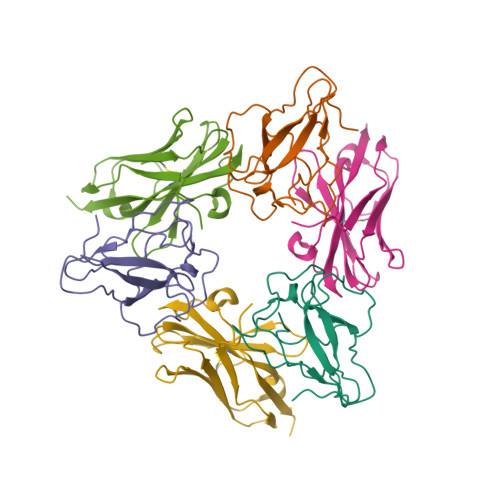
RCSB PDB - 7R98: Structure of the SARS-CoV-2 N protein RNA-binding domain bound to single-domain antibody B6
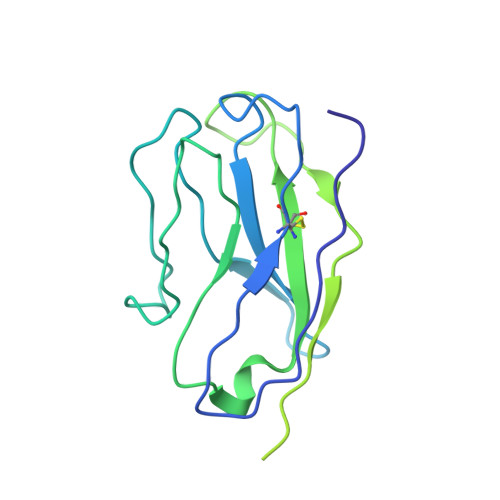
RCSB PDB - 7R40: Structure of the SARS-CoV-2 spike glycoprotein in complex with the 87G7 antibody Fab fragment

RCSB PDB - 1R7C: NMR structure of the membrane anchor domain (1-31) of the nonstructural protein 5A (NS5A) of hepatitis C virus (Minimized average structure, Sample in 50% tfe)
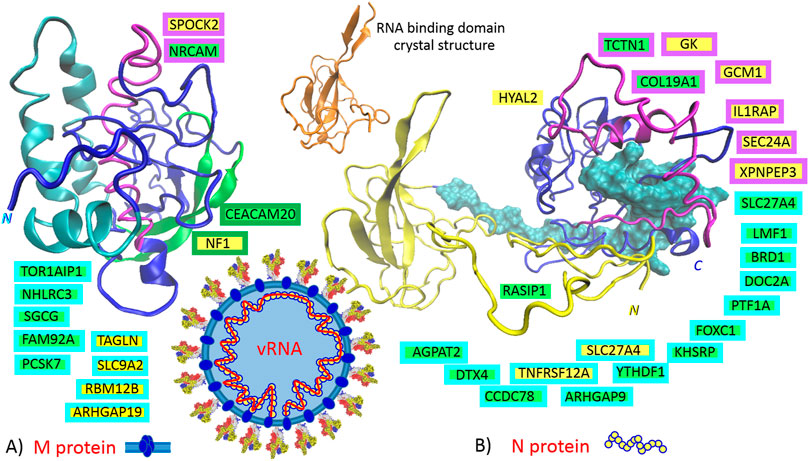
Frontiers Theoretical Analysis of S, M and N Structural Proteins by the Protein–RNA Recognition Code Leads to Genes/proteins that Are Relevant to the SARS-CoV-2 Life Cycle and Pathogenesis
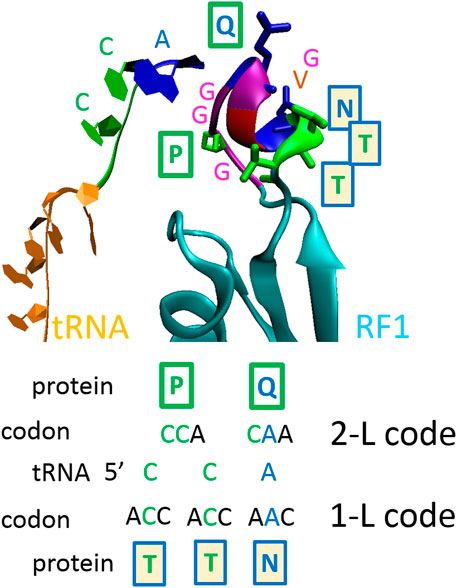
Frontiers Theoretical Analysis of S, M and N Structural Proteins by the Protein–RNA Recognition Code Leads to Genes/proteins that Are Relevant to the SARS-CoV-2 Life Cycle and Pathogenesis
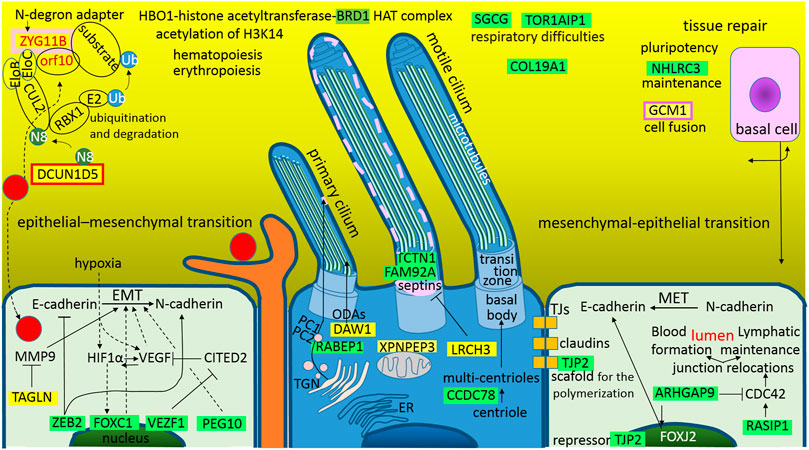
Frontiers Theoretical Analysis of S, M and N Structural Proteins by the Protein–RNA Recognition Code Leads to Genes/proteins that Are Relevant to the SARS-CoV-2 Life Cycle and Pathogenesis
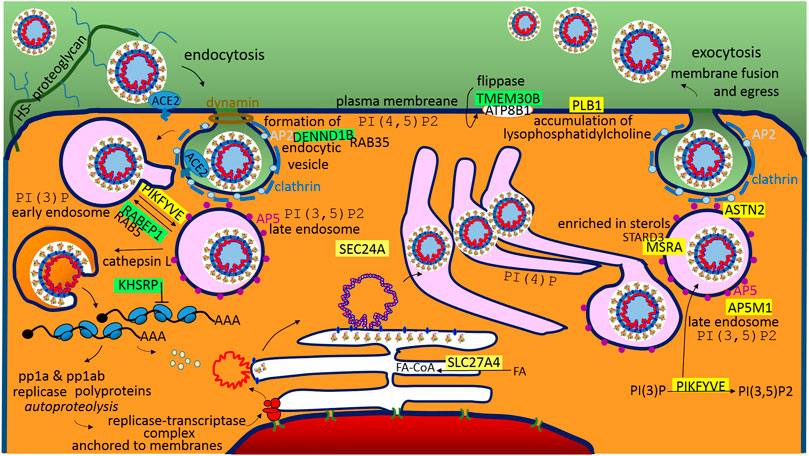
Frontiers Theoretical Analysis of S, M and N Structural Proteins by the Protein–RNA Recognition Code Leads to Genes/proteins that Are Relevant to the SARS-CoV-2 Life Cycle and Pathogenesis

RCSB PDB - 7R98: Structure of the SARS-CoV-2 N protein RNA-binding domain bound to single-domain antibody B6

The preference signature of the SARS-CoV-2 Nucleocapsid NTD for its 5'-genomic RNA elements
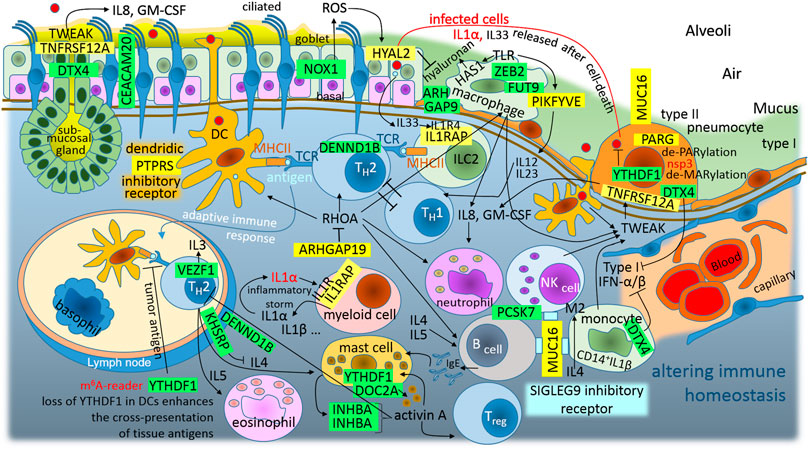
Frontiers Theoretical Analysis of S, M and N Structural Proteins by the Protein–RNA Recognition Code Leads to Genes/proteins that Are Relevant to the SARS-CoV-2 Life Cycle and Pathogenesis
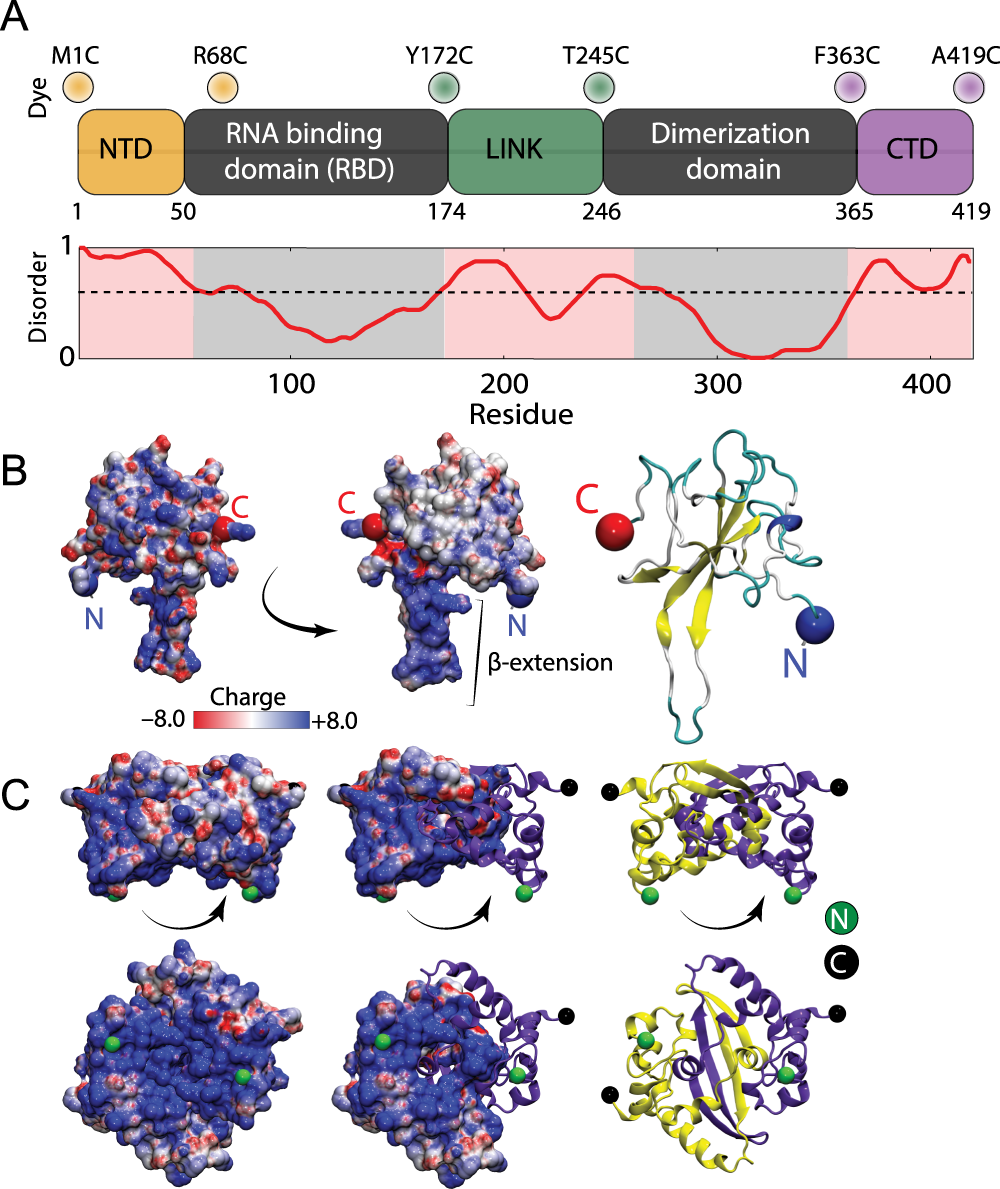
The SARS-CoV-2 nucleocapsid protein is dynamic, disordered, and phase separates with RNA